FIGURE
1 (caption below)This image may be
enlarged
in edit mode of your browser (eg: composer, Frontpage)
Online Journal of Veterinary Research
Volume 1:1-6, 2002
Detection of sheep-goat interspecies hybridization by analysis of satellite DNA
Isaäc J. Nijmana, Martinus P. Hogendoornb, Erik Gruys a, Gordon Luikartc, Okan Ertugruld, Yo Zagdsuren e, L.O. Ngeref, Noor Hasimag, Georg Erhardt h, Paolo Ajmone-Marsani and Johannes A. Lenstraa
a Faculty of
Veterinary
Medicine, Utrecht University Yalelaan 1, 3584 CL Utrecht, The
Netherlands
b Delftsestraatweg 14, 2641 NB Pijnacker, The Netherlands c Laboratoire
de Biologie des Populations d'Altitude, Centre National de la Recherche
Scientifique, Unité Mixte de Recherche 5553, Université
Joseph
Fourier, B.P. 53, F-38041 Grenoble Cedex 9, France d Ankara
Universitesi
Veteriner Fakültesi, 06110 Diskapi, Ankara, Turkey, e Research
Institute
of Animal Husbandry, Zaisan 210153, Ulaanbaatar, Mongolia f Dept.
Animal
Science, University of Ibaden, Ibaden, Nigeria g Dept. Animal Genetics
and Cellular Biology, Institute Science Biology, University of Malaya,
50603 Kuala Lumpur, Malaysia h Dept. Animal Breeding and Genetics,
Justus-Liebig-Universität,
Ludwigstrasse 21 B, D-35390 Giessen, Germany i Institute of
Zootechnics,
Catholic University of S. Cuore, via Emilia Parmense, 84, I-29100
Piacenza,
Italy.E
Email: J.A.Lenstra@vet.uu.nl
Nijman IJ, Hogendoorn MP, Gruys E, Luikart G, Ertugrul O, Zagdsuren Y, Ngere LO, Hasima N, Erhardt G, Ajmone-Marsan P, Lenstra JA. Detection of sheep-goat interspecies hybridization by analysis of satellite DNA, Online Journal of Veterinary Research 1:1-6, 2002. Sequence analysis of sheep and goat satellite II allowed the identification of species-specific mutations that can be detected conveniently by satellite restriction fragment length polymorphism (SFLP) analysis. Restriction enzyme patterns did not depend on the sheep or goat breed that were analyzed, while also ibex and bezoar, two wild goat species, had the same patterns as domestic goat. The satellite II SFLP provides a rapid assay of sheep-goat hybridization, which allowed the verification and disapproval, respectively, of two cases of suspected goat-sheep hybridization.
KEYWORDS:
Sheep, Goat, Hybrids, Species Origin, Satellite DNA, PCR
Sheep (Ovis aries ) and goat (Capra hircus) both belong to the bovid subfamily of the Caprinae and are brought into contact by their common domestication. Interspecies mating occurs occasionally and ram doe matings may result in pregnancy (Kelk et al 1997 ). Usually, hybrid fetuses die within 5 to 10 weeks, but the birth of live animals has been reported (Tucker et al 1989 ; Gustafson et al 1993 ; Letshwenyo & Kedikilwe, 2000 ). Fertilization of ewes by bucks in the field, often suspected on the basis of a deviating appearance of the lamb, has never been verified ( McGovern, 1969 ; Gray, 1971 ; Long, 1990 ). Hybrid pregnancy of ewes can be established by artificial insemination and embryo transfer, but ends after 4 to 6 weeks of pregnancy ( Gustafson et al 1993 ). In general, sheep-goat hybridization is a suitable model for studying the reproductive barrier between species (Anderson 1988 ; Roth 1989 ; Oppenheim et al 2000).
Several methods are available for verification of an interspecies origin, such as karyotyping ( Hancock and Jacobs 1966; Märki and Osterhoff 1984 ; Denis et al 1987 ), blood protein typing (Tucker et al 1989 ), or parentage testing by microsatellite genotyping. However, these methods are too complicated for occasional sporadic use. Consequently, suspected sheep-goat hybridization is only rarely confirmed by scientific evidence ( Märki and Osterhoff 1984; Tucker et al 1989 ; Letshwenyo and Kedikilwe 2000 ).
Satellite DNA is organized as a tandem repeat in the centromeric regions of the chromosomes and accounts for up to 20 percent of the genome. Previously, we described the analysis of the satellite DNA in order to detect introgression of zebu in African taurine cattle (Nijman et al 1999 ) or hybridization of zebu and banteng in Indonesia (Nijman 1999 ). Unlike mitochondrial DNA, the nuclear satellite DNA is transmitted via both parental lines. Satellites of related species usually differ by mutations in the repeat units (Jobse et al 1995 ; Nijman 1999 ). Mutations that create or abolish a restriction site can be tested by SFLP (satellite fragment length polymorphism, Nijman et al 1999). Here we report the sequences of amplified goat and sheep satellite II and identify sheep- and goat-specific restriction sites, which do not depend on the breed and are also shared by related wild species. This allows a rapid test for verification of presumed sheep-goat hybridizations.
MATERIALS AND METHODS
DNA isolation: Case 1: DNA was isolated from necrotic tissue from a 93 d-old fetus by treatment with SDS and proteinase K and phenol extraction (Sambrook et al 1989 ). Case 2: heparinized blood was collected from a sheep lamb suspected to be sired by a billy goat. DNA was isolated from lymphocytes by extraction with guanidinium isothiocyanate as described by Ciulla et al (1988 ). Blood samples from Rhön sheep, Graue Gehörnte Heidschnucke, Romanov and Weisses Bergschaf were collected at the Research Station Oberer Hardthof of the Justus Liebig University in Giessen. DNA was isolated from sheep lymphocytes as described previously ( Montgomery and Sise 1990 ). DNA was isolated from skin biopsies and blood from Angora, Mongolian Cashmer, Bayendelger Casmer, West-African Dwarf, Malaysian native, goats, from a Mongalian ibex (Capra ibex sibirica) and from a Caucasian bezoar (Capra aegagrus ) by using the Qiagen kits for tissue and blood, following the manufacturer’s instructions. DNA was isolated from bloods samples of Sarda and Bionda dell’Adamollo goats by CTAB extraction (Saghai-Maroof et al 1984 ).
PCR and sequencing: Primers 5'-GTTGCACATCCAAGGGCTCC and 5'-CCGGGCAGAGCAGCCTCG were based on cloned sequences of goat and sheep satellite II (Buckland, 1985; Burkin et al 1996) and directed the amplification of a 535 bp fragment from the 705-bp repeat unit. PCR was performed in 50 µl containing 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris/HCl (pH 8.3), 0.01% gelatine, 0.2 mM NTPs, 50 ng genomic DNA from sheep, goat or suspected hybrids, 50 ng of both primers, 1.25 U Taq DNA polymerase (Promega). The following program was used in an ABI 9600 thermocycler: 2 min at 92°C, 30 cycles of 15 s at 92°C, 45 s at 60°C, and 45 s at 72°C. A 528-bp DNA fragment was isolated by electrophoresis in a 2% agarose/TBE gel (Sambrook et al 1989) and Qiaquick (Qiagen) purification. PCR products were sequenced by extension of 3 pmol primer in a 10 µl reaction volume containing 5 to 20 ng PCR product and the components of the ABI BigDye terminator cycle sequencing kit and by fractionation of extension products on an ABI 310 fluorescent automatic sequencer with fluorescent dideoxy nucleotides. The sheep and goat sequences have been deposited in the Genbank database with accession numbers AF245169 and AF245170, respectively.
A 359-bp cytochrome b fragment from Capra ibex siberica was amplified as described previously ( Meyer et al 1995; Branciari et al 2000 ) and sequenced as described above. Data are accessible via the Genbank accession code AF424636.
Sheep/goat species test: After amplification of the 528-bp segment from the repeat unit of satellite II as described above, the PCR product (10 µl) was digested by adding the prescribed concentrated reaction buffer, 5 to 10 U RsaI or TaqI (Amersham Pharmacia Biotech) and H 2O to 15 µl. After incubation for 2 h at 37°C (RsaI) or 65°C (TaqI), respectively, digestion products were analyzed on a 2% agarose/TBE gel (Sambrook et al 1989 ).
RESULTS
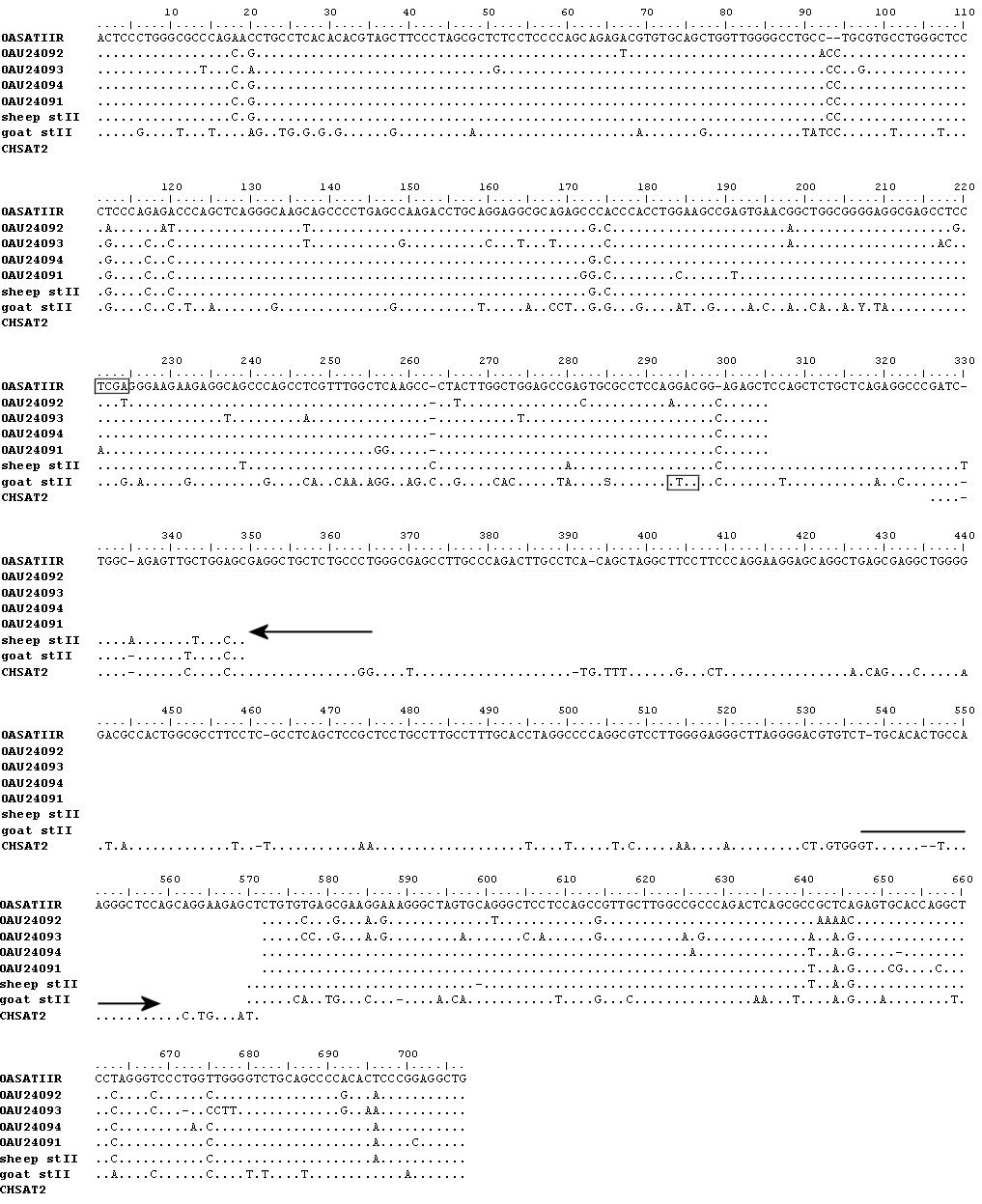
PCR primers were based on an alignment of sequences of cloned satellite II from sheep ( Buckland, 1985 ; Burkin et al 1996) and a partial sequence from goat (Buckland, 1985 ). The sequences of the resulting PCR products have been aligned with the published data (Fig. 1). The sheep PCR product represents the several repeat units that were amplified simultaneously and its sequence agrees well with the cloned sequences of single repeat units. The sequence of the goat PCR product has only a small overlap with the published partial sequence and differs considerably from the sheep sequence.
FIGURE
1 (caption below)This image may be
enlarged
in edit mode of your browser (eg: composer, Frontpage)

Figure 1. Alignment of goat and sheep satellite II sequences. OASATIIR and OAU24091 to OAU24094 are cloned sequences from sheep (Buckland, 1985 ; Burkin et al 1996 ) and CHSAT2 is a cloned sequence from goat (Buckland, 1985). Dots indicate identity to the OASTAIIR sequences and dashes indicate the positions of deletions or insertions. ‘Sheep stII’ and ‘goat stII’ designate newly determined sequences of PCR products from sheep and goat, respectively. PCR primers for generating these products and for the species test are indicated by arrows. The sheep-specific TaqI site and the goat-specific Rsa I site are boxed. This image may be enlarged in edit mode of your browser (eg: composer, Frontpage)
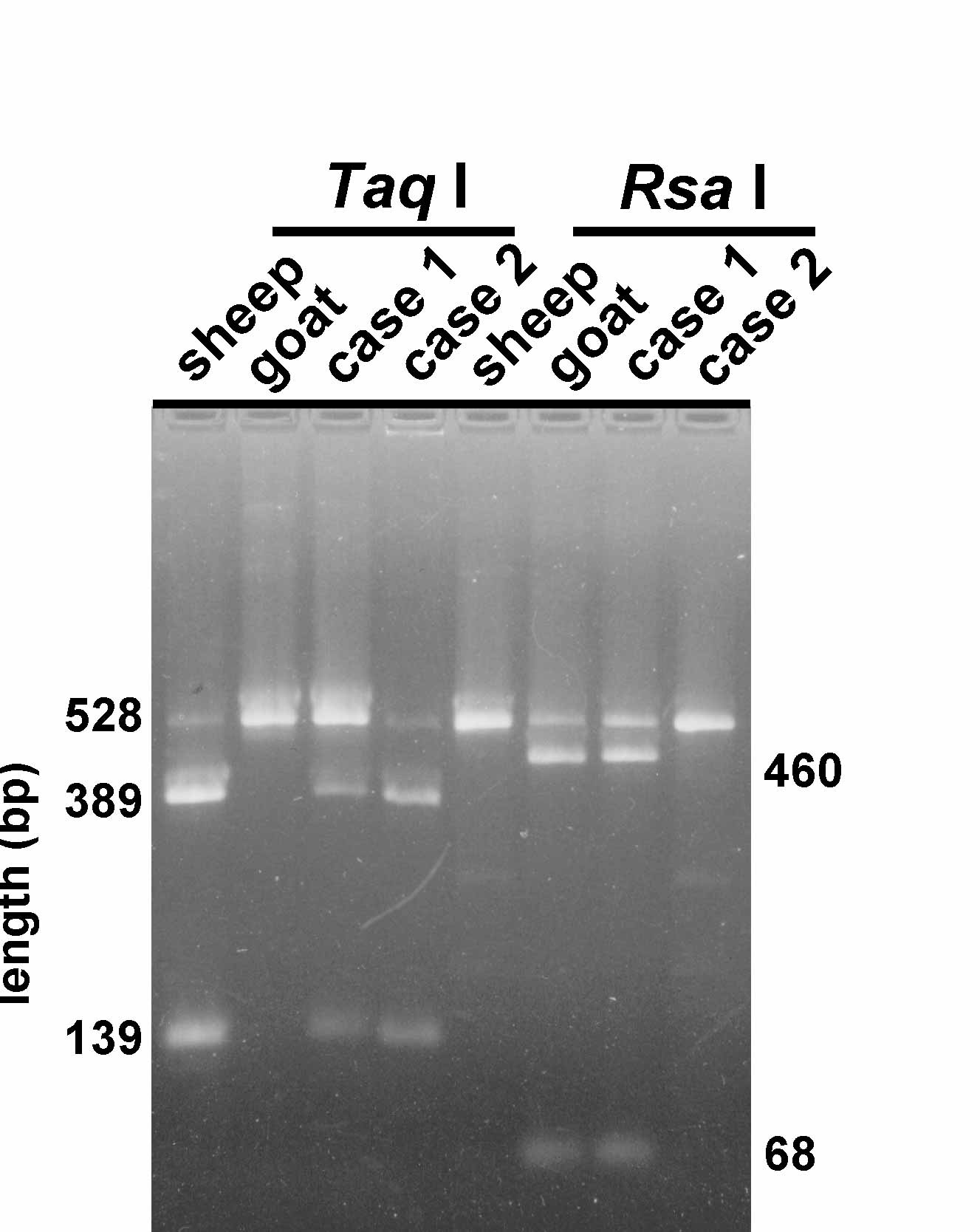
Figure 2 . Restriction digestions of sheep and goat satellite II. The lengths of the intact PCR product (528 bp) and the digestion products are indicated. Case 1 was a fetus from a goat after a pregnancy of 93 days. Case 2 was a sheep lamb suspected to have been sired by a goat.
A search for restriction-enzyme recognition sequences revealed a sheep-specific TaqI site and a goat-specific RsaI site. Figure 2 shows that these enzymes indeed generated species-specific SFLP profiles. As indicated by the degree of digestion, the TaqI is present in almost all sheep satellite II repeat units, whereas the RsaI site is in most goat units. SFLP analysis of satellite II from various sheep breeds, goat breeds (see the Materials and Methods), the related goat species ibex (Capra ibex sibericus ) and bezoar (Capra aegagrus) and digestion with the same restriction enzymes showed that the patterns were indeed specific for sheep and goat, respectively (results not shown).
Two field cases studied in this paper were a fetus from a goat fertilized by a ram (case 1) and a sheep lamb, which was considered goat-like in appearance and behavior after the presence of a billy goat during the mating period (case 2). The hybrid origin of case 1, the stillborn fetus from a goat, is clearly indicated by the Taq I pattern (Figure 2). However, the suspected hybrid origin of case 2, a lamb born from a sheep, could be disproved on the basis of both the Taq I and the RsaI patterns.
DISCUSSION
Variation in DNA sequence provides several categories of markers for the elucidation of genetic or phylogenetic relationships (Nijman, 1999 ): mutations in mitochondrial or nuclear sequences, insertion of SINE or LINE elements, length variation of mini- and microsatellites, and variation of centromeric satellite DNA. Nowadays, for most markers the detection of alleles or species-specific variants is based on a convenient and sensitive PCR assay. The choice of the marker most appropriate for a specific application is dictated by an appropriate level of variability at the required phylogenetic distance.
Detection of
sequence
variants of satellite DNA has, relative to other methods, the following
advantages for the identification of species hybrids:
1. The chemical stability of DNA allows the analysis of dead and even historic tissue and does not require, like karyotyping, the culturing of live cells (Märki and Osterhoff, 1984; Long, 1990 ).
2. No DNA material of the sire is required. This is an advantage relative to parentage testing if the sire has been slaughtered before the birth of the offspring or is not available for other reasons.
3. The genome contains many copies of the satellite DNA repeat unit, which makes their PCR amplification easier than the amplification of single-copy genes or microsatellites. This is an advantage for the analysis of hair samples, necrotic tissue, historic material, etc.
4. Satellite DNA is transmitted via both parental lines, so individuals will contain species-specific variants from both the father and the mother. This is in contrast to the maternal transmission of mitochondrial DNA, which is a sensitive marker for the origin of food products from sheep and goat (Meyer et al 1995 ; Branciari et al 2000 ), but is not informative for the identification of the species of the sire.
5. Species-specific mutations in satellite DNA often create or abolish the recognition sequence of a restriction site, which allows the detection of these mutations by PCR, restriction digestion and agarose gel electrophoresis (Jobse et al 1995; Nijman et al 1999 ). This does not require radioactive or fluorescent labeling like microsatellite analysis.
6. Generally, the resulting SFLP pattern does not depend on the breed (Nijman et al 1999 , this paper). We also observed that the SFLP patterns of the wild goat species bezoar and ibex were identical to the pattern of domestic goat, showing that the frequency of the restriction site has remained stable during the domestication period. In contrast, restriction sites in the cytochrome b gene, commonly used for species identification (Meyer et al., 1995), were different in domestic and wild goats (Wolf et al 2000; Genbank accession AF424636).Chikuni et al (1994) and Jobse et al (1995 ) described digestions by ApaI and EcoRI, respectively, of sheep and goat satellite I PCR products. In sheep a large proportion of the satellite I repeat units has an ApaI as well as an EcoRI site, which are both absent in goat satellite I but will be present in a sheep-goat hybrid. Indeed, this allowed the verification of the hybrid origin of case 1, a fetus resulting from a ram x doe mating (not shown). In this case the maternal goat component needed no verification and the digestion products unambiguously identified a ram as father. However, satellite I appeared not suitable for analysis of the reciprocal buck x ewe mating for two reasons. First, no goat-specific restriction sites could be identified, so the difference between the patterns of sheep and a hybrid would only be a quantitative shift. Second, in mixtures of sheep and goat DNA the amplification of the sheep satellite I appeared to predominate, further masking the difference between pure sheep and hybrid lambs
Therefore, we
determined
the sequence of PCR products of goat and sheep satellite II, which is
homologous
to the bovine satellite II and accounts for 2.5 % of the sheep and goat
genome, respectively (Kurnit
et al 1978 ). Diagnostic sites for both the ovine and caprine
sequences
were identified. No preferential amplification of either the goat or
the
sheep satellite II was observed in mixtures of goat and sheep DNA.
Testing
of two cases of suspected cases of sheep-goat hybridization led to a
verification
and a disapproval, respectively. Case 2 was a typical case of suspected
hybridization, which in most cases is never tested. Our test requires
only
a standard PCR reaction, digestion by restriction enzymes and agarose
gel
electrophoresis, but does not need access to expensive DNA genotyping
methodology.
Acknowledgments:
We thank the late Mr. G. Weerheim (Pijnacker, The Netherlands) and Mr. P.J. Smit (Schipluiden, The Netherlands) for access to suspected sheep-goat hybrids.
REFERENCES
Branciari RS, Nijman IJ, Plas ME, Di Antonio E., Lenstra JA (2000), Species origin of milk in Italian mozzarella cheese and Greek feta, Journal of Food Protection 63, p 408-411
Buckland RA (1985), Sequence and evolution of related bovine and caprine satellite DNAs. Identification of short DNA sequence potentially involved in satellite DNA amplification, Journal of Molecular Biology 186 p 25-30
Burkin DJ, Broad TE, Jones, J. (1996), The chromosomal distribution and organization of sheep satellite I and II centromeric DNA using characterized sheep-hamster somatic cell hybrids, Chromosome Research 4 p 49-55
Chikuni K, Tabata T, Kosugiyama M, Monma M, Saito M (1994), Polymerase chain reaction assay for the detection of sheep and goat meats, Meat Science 37 p 337-345
Ciulla TA, Sklar SM, Hauser SL (1988), A simple method for DNA purification from peripheral blood, Analytical Biochemistry 174 p 485-488
Denis B, Malher X, Seegers H, Darre R, Berland H (1987), Note sur et à propos d’un hybride chèvre x mouton fertile. Elevage et Insemination 219 p 3-10
Gray AP (1971), Mammalian hybrids. Commonwealth Agricultural Bureaux, Edinburgh, UK
Gustafson RA, Anderson GB, BonDurant RH, Sasser GR (1993). Failure of sheep-goat hybrid conceptuses to develop to term in sheep-goat chimaeras. Journal of Reproduction and Fertility 99 p 267-273
Hancock JL, Jacobs, PA (1966), The chromosomes of goat x sheep hybrids. Journal of Reproduction and Fertility 12 p 591-592
Jobse C, Buntjer JB, Haagsma N, Breukelman HJ, Beintema JJ, Lenstra JA (1995), Evolution and recombination of repetitive DNA from the ruminants. Journal of Molecular Evolution 41 p 277-283
Kelk DA, Gartley CA, Buckrell BC, King WA (1997), The interbreeding of sheep and goats. Canadian Veterinary Journal 38 p 235-237
Kurnit, DM, Brown FL, Maio JJ (1978) Mammalian repetitive DNA sequences in a stable Robertsonian system. Characterization, in situ hybridizations, and cross-species hybridizations of repetitive DNAs in calf, sheep, and goat chromosomes. Cytogenetics and Cell Genetics 21 p 145-167
Letswenyo M, Kedikilwe K (2000), Goat-sheep hybrid born under natural conditions in Botswana. Veterinary Record 146 p 732-734
Long, SE (1990), Chromosomes of sheep and goat. Advances in Veterinary Science and Comparative Medicine 34 p109-129
Märki U, Osterhoff DR (1984), Disproval of an apparent goat x sheep hybrid. Journal of the South African Veterinary Assocication 55 p 133-134
Montgomery GW, Sise JA (1990), Extraction of DNA from sheep white blood clees. New Zealand Agricultural Research 33 p 437-441
Nijman IJ, Bradley DG, Hanotte O, Otsen M, Lenstra JA (1999), Satellite DNA length polymorphism and AFLP correlate with Bos indicus-taurus hybridization. Animal Genetics 30 p 265-273
Nijman IJ (1999), Repetive DNA elements as genetic and phylogenetic markers in the genomes of cattle and other ruminants. Ph.D. dissertation, Utrecht University, Utrecht
Oppenheim SM, Moyer AL, BonDurant RH, Rowe, JD, Anderson GB (2001), Evidence against humoral immune attack as the cause of sheep-goat interspecies and hybrid pregnancy failure in the doe, Theriogenology 55 p 1567-1581
Roth TL, Anderson GB, Bon Durant RH, Pashen RL (1989), Survival of sheep x goat inner cell masses after injection into ovine embryos. Biology of reproduction 41 p 675-682
Saghai Maroof SA, Soliman KM, Jorgensen RA, Allard RW (1984), Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences USA, 81 p 8014-9018
Sambrook J, Fritsch EF, Maniatis T (1989), Molecular Cloning. A Laboratory Manual (2nd Ed.), Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Tucker EM, Denis B, Kilgour L (1989), Blood genetic markers studies of a sheep-goat hybrid and its back-cross offspring. Animal Genetics 20 p 170-186